Timisoara_Med 2021, 2020(2), 6; doi:10.35995/tmj20200206
Article
Long Non-Coding RNAs in Plasma and Urine as Potential Biomarkers in Prostate Cancer
1
Department of Urology, Victor Babeș University of Medicine and Pharmacy, 300041 Timișoara, Romania; paunescuionut@yahoo.com (I.A.P.); alincumpanas@hotmail.com (A.C.)
2
Department of Urology, Prof. Dr. Ion Chiricuță Oncological Institute, 400015 Cluj-Napoca, Romania; uropraxb@gmail.com
3
Department of Functional Genomics and Experimental Pathology, Prof. Dr. Ion Chiricuță Oncological Institute, 400015 Cluj-Napoca, Romania; obalacescu@yahoo.com
4
Department of Pathology, Victor Babeș University of Medicine and Pharmacy, 300041 Timișoara, Romania; dema_alis@yahoo.com
5
Department of Biochemistry, Victor Babeș University of Medicine and Pharmacy, 300041 Timișoara, Romania; marcu.anca@umft.ro (A.M.); cmarian@umft.ro (C.M); eseclaman@umft.ro (E.S.); ovidiu.sirbu@umft.ro (O.S.)
*
Correspondence: razvan.bardan@gmail.com; Tel.: +40-723-307-888
How to cite: Păunescu, I.A.; Bardan, R.; Petruț, B.; Bălăcescu, O.; Cumpănaș, A.; Dema, A.; Marcu, A.; Marian, C.; Șeclăman, E.; Sîrbu, I.O. Long Non-Coding RNAs in Plasma and Urine as Potential Biomarkers in Prostate Cancer. Timisoara Med. 2020, 2020(2), 6; doi:10.35995/tmj20200206.
Received: 17 January 2021 / Accepted: 3 February 2021 / Published: 8 February 2021
Abstract
:(1) Introduction: Prostate cancer is the second leading cause of cancer-related death in men in developed countries. Due to the existing biomarkers’ limitations, there is a stringent need to develop novel, better non-invasive markers for prostate cancer diagnostic and monitoring. (2) Material and methods: We assessed, by real-time PCR, the expression level of 84 long non-coding RNA (lncRNA) in plasma and the exosomes isolated from prostate cancer patients’ plasma and urine. (3) Results: Only a few lncRNAs were detected in high abundance (Ct between 25 and 30 cycles) across all sample types, the vast majority showing relatively modest levels (Ct > 30 cycles). As expected, plasma and plasma exosomes contain far more lncRNA species than urine, irrespective of whether they originate from patients or controls. We identified two statistically significant dysregulated lncRNAs in prostate cancer samples vs. controls: RBM5-AS1, 2.89 times downregulated in plasma (p = 0.036), and SNHG16, 13.69 times upregulated (p = 0.029) in urine exosomes. (4) Conclusions: These preliminary data need further validation in additional independent, more extensive studies before they can be considered as biomarkers for prostate cancer.
Keywords:
prostate cancer; biomarker; lncRNAIntroduction
Prostate cancer is the second leading cause of cancer-related death in men in developed countries. Significant efforts are still needed for its early diagnosis and monitoring using both clinical and laboratory tests [1]. The current, blood-based biomarkers rely mainly on protein testing, are labor-intensive and expensive, and have modest sensitivity and specificity [2]. There is an unmet need for further research into better biomarkers for prostate cancer detection and monitoring.
Less than 3% of the human genome consists of protein-coding genes, while more than 80% is actively transcribed into RNA without being translated into proteins, the so-called non-coding RNAs, which are arbitrarily classified into small (<200 nucleotides long) and long (>200 nucleotides long) ncRNA [3]. Small ncRNAs (microRNAs in particular) have been extensively studied in relation to various pathologies, including cancer; however, much less is known about the function and biomarker potential of the lncRNAs. Nevertheless, there is increasing evidence regarding their association with normal and pathological conditions, including cancer [4].
Multiple studies have investigated the potential role of lncRNAs as biomarkers in prostate cancer (reviewed in Shukla et al., 2020); most of them have detected lncRNA in urine, while only a few studies have comparatively investigated both urine and plasma [5]. The role of prostasomes (microvesicles secreted by prostate cells, including prostate cancer cells) as carriers of biomarkers for prostate cancer has long been investigated; however, only recently has it been shown that they carry multiple types of RNA, including lncRNAs [6,7].
Herein, we have investigated the biomarker potential of 84 lncRNAs from plasma and exosomes purified from plasm and urine in prostate cancer patients and cancer-free controls.
Material and Methods
We collected plasma and urine samples from 14 patients with prostate cancer and 15 cancer-free controls diagnosed and treated in the Urology Clinic of the Clinical Emergency County Hospital in Timisoara, Romania. All subjects provided informed consent for use of their biological samples in the present study; the study was approved by the Ethics Committees of the participating institutions (the Clinical Emergency County Hospital in Timisoara and the Victor Babes University of Medicine and Pharmacy Timisoara).
Prostate-specific antigen (PSA) testing was performed for both cancer patients and healthy controls in the Laboratory of the Timisoara Clinical Emergency County Hospital, using the Abbott Diagnostics (Chicago, IL, USA) chemiluminescent microparticle immunoassay (CMIA).
Venous blood was collected in ethylenediaminetetraacetic acid (EDTA)-treated blood collection tubes and was immediately centrifuged for 15 min at 2000 g for plasma separation and frozen at −80 °C until further use. Urine samples were collected from patients and controls in 50 mL sterile collection tubes, centrifuged for 10 min at 1000 g to pellet cells and debris, and the supernatant was stored at −80 °C until further use. The plasma and urine lots (control and prostate cancer) were built by pooling 100 μL individual plasma and 250 μL individual urine samples, respectively.
The pooled plasma samples were subject to total RNA extraction using either miRNeasy Serum/Plasma kit (Qiagen, Hilden, Germany) or Total Exosome RNA and Protein Isolation Kit (ThermoFisher, Waltham, MA, USA). Each procedure was performed in triplicate, starting from 200 μL pooled plasma. Urine exosomes were purified using Total Exosome Isolation (from urine) (ThermoFisher, Waltham, MA, USA), starting from 1 mL pooled urine. The total RNA was purified from urine exosome using Total Exosome RNA and Protein Isolation Kit (ThermoFisher, Waltham, MA, USA). The RNA concentration was assessed with Qubit 2.0 (ThermoFisher, Waltham, MA, USA), using the Qubit RNA BR (broad range) Assay Kits.
From each extracted pooled sample, the same amount of total RNA (10 ng) was reverse-transcribed using RT2 First Strand kit (Qiagen, Hilden, Germany), according to the manufacturer’s indication. Simultaneous expression analysis of lncRNAs was performed on a 7900 HT Real-Time PCR System (ThermoFisher) using RT2 lncRNA PCR Array Human lncFinder (Qiagen, Hilden, Germany). Each real-time experiment was performed in triplicate.
Data analysis was performed according to the ΔΔCt method with a Ct cutoff value of 39, using Qiagen’s GeneGlobe Data Analysis Center platform. The most suitable normalization method was used for each sample type, as selected by the algorithm in the analysis platform: RPLPO housekeeping gene for plasma samples, the average geometric mean of PANDAR lncRNA and ACTB housekeeping gene for plasma exosomes samples, and average geometric mean of housekeeping genes ACTB, RPLP0, RN7SK, SNORA73A for urine exosome samples.
Results
The clinical and demographic characteristics of the study subjects are presented in Table 1.
On average, control subjects were younger than patients and had lower serum PSA values than prostate cancer patients, with mean PSA values of 0.98 ng/mL and 17.27 ng/mL, respectively. Of note, the Gleason score for most (n = 12, 85.71%) of the cancer patients was above 6.
Table 2 presents the proportion of the targets detected in our samples, as percent of the entire Human lncFinder RT2 lncRNA PCR Array plate. While the housekeeping genes and the controls assessing reverse transcription and PCR performance were detected early on during the PCR process (Ct < 25), just a few lncRNAs were found to be abundantly expressed (25 < Ct < 30), with the vast majority of lncRNAs being detected after Ct = 30.
Regarding the distribution of detected lncRNAs across sample types, Figure 1 presents Venn diagrams of the expressed lncRNAs in controls (left) and cancer cases (right). More lncRNAs were detected in plasma samples (both total plasma and exosomes) compared to urine, both in patients and controls. We identified one single lncRNA in urine exosomes from controls and five lncRNAs in urine exosomes from patients. Among lncRNAs detected in plasma, only one was present exclusively in the exosomes from controls’ plasma, and six in the exosomes from patients’ plasma.
When comparing cases to controls, we set a threshold of two-fold difference for differentially expressed lncRNAs. All differentially expressed lncRNAs are presented in Table 3.
In plasma samples, we found RBM5-AS1 to be the only lncRNA that was statistically significantly dysregulated (p = 0.036), being 2.89 times under-expressed in prostate cancer samples compared to controls (Figure 2). No lncRNA was detected as being differentially expressed in the exosomes purified from plasma of prostate cancer patients, compared to controls at the significance value of p = 0.05. In exosomes extracted from urine, we found one lncRNA (SNHG16) to be statistically significantly dysregulated (p = 0.029), being 13.69 times over-expressed in prostate cancer samples compared to controls (Figure 3).
Discussion
Our data identify RBM5-AS1 and SNHG16 as novel putative prostate cancer biomarkers. RNA Binding Motif Protein 5 Antisense RNA 1 (RBM5-AS1) is 1387 nucleotides long, transcribed from an otherwise banal locus on the chromosome 3 (chr3:50,099,602-50,100,988; GRCh38/hg38), where it overlaps with RBM5, NONHSAG035102.2, and partially with RBM6 and. To our knowledge, this is the first description of RBM5-AS1 as a putative biomarker associated with prostate cancer. S, o date-AS1 has been linked only to the oral squamous cell carcinoma, where it promotes the proliferation, migration, and invasion behavior of OSCC through miR-1285-3p/YAP1 axis, and to colon cancer, where it is involved in the self-renewal of cancer stem cells [8,9].
Small Nucleolar RNA Host Gene 16 (SNHG16) is a lncRNA commonly known for its association with neuroblastoma and bladder, transcribed from chr17:76,557,359-76,792,366 (GRCh38/hg38), a region rich in long and small non-coding RNAs. SNHG16 is a somewhat promiscuous malignancy marker, as it has been associated with a wide array of malignancies in humans (cervical cancer, esophageal cancer, gastric cancer, etc.). SNHG16 has been proposed not only as a biomarker but, interestingly, also as a potential therapeutic target [10]. Recently, SNHG16 was shown to inhibit prostate cancer cell growth ex vivo, a mechanism allegedly mediated by GLUT-1 [11].
The number of human lncRNAs has steadily increased as the human genome is annotated, and, despite the somewhat limited information on their function, there is increasing evidence outlining their involvement in multiple biological processes in normal and pathological conditions. It has been shown that lncRNAs are very specific for certain cancers, including prostate cancer, and given that they outnumber the protein-coding genes, it is highly likely that the next prostate cancer biomarker will be a non-coding RNA.
Transcriptomic profiling of prostate cancer tissue has identified over seven hundred lncRNAs, including PCA3, SChLAP1, NEAT1, PCAT1, PCAT14, PVT1, PCAN-R1, and PCAN-R2 [12,13]. Some of these lncRNAs have been detected in the circulation and/or urine and are thus exciting candidates as minimally invasive markers next to Prostate Cancer Antigen 3 (PCA3), the first known non-coding RNA with biomarker qualities [14,15]. PCA3 is 34-fold more highly expressed in prostate cancers than normal tissue and can be detected in the urine sediment, aiding (in conjunction with PSA) in selecting the ambiguous cases that would benefit from a prostate biopsy [16].
SChLAP1 is overexpressed in aggressive prostate cancers and was detected in the urine sediment of patients at risk of developing metastatic progression [17,18]. Urinary MALAT1 has been linked to prostate cancer, especially in subjects with PSA levels between 4 and 10 ng/mL [19]. Urinary exosomal lncRNA-p21 is of particular interest since it harbors miR seed regions and has an excellent profile upon evaluation in preclinical settings [20]. Certain urinary lncRNAs have been proposed as therapy response predictors: the estrogen-sensitive NEAT1 has been associated with resistance to anti-androgen therapy and PCAT1 confers sensitivity to PARP1 inhibitors [21].
Conclusions
In this pilot project, we found several lncRNAs that were differentially expressed in plasma and exosomes isolated from plasma and urine of prostate cancer patients compared to normal controls, although only two of these markers reached statistical significance. These preliminary data need further confirmation and validation in additional, independent, more extensive studies before they can be considered as biomarkers for prostate cancer.
Author Contributions
conceptualization: I.A.P., R.B., and C.M.; methodology: I.A.P., B.P., O.B., A.M., and E.S.; validation: A.D., A.M., and C.M.; formal analysis: O.B., A.M., and C.M.; investigation: I.A.P., B.P., O.B., A.M., E.S., and I.O.S.; writing—draft preparation: I.A.P., R.B., A.C., and C.M.; writing—review and editing: I.A.P., R.B., B.P., A.C., C.M., and I.O.S.; supervision: R.B., and C.M., project administration: C.M. and E.S., funding acquisition: C.M.
Funding
This research was funded by UEFISCDI, grant PN-III-P4-ID-PCE-2016-0371.
Acknowledgments
We are grateful for Mirela Mititelu’s technical support.
Conflicts of Interest
The authors declare no conflict of interest.
References
- American Cancer Society. Cancer Facts & Figures 2016; American Cancer Society: Atlanta, GA, USA, 2016. [Google Scholar]
- Scatena, R. Advances in Cancer Biomarkers; Springer Science + Business Media: Dordrecht, The Netherlands, 2015. [Google Scholar]
- Martens-Uzunova, E.S.; Böttcher, R.; Croce, C.M.; Jenster, G.; Visakorpi, T.; Calin, G.A. Long Noncoding RNA in Prostate, Bladder, and Kidney Cancer. Eur. Urol. 2014, 65, 1140–1151. [Google Scholar] [CrossRef] [PubMed]
- Ramalho-Carvalho, J.; Fromm, B.; Henrique, R.; Jerónimo, C. Deciphering the function of non-coding RNAs in prostate cancer. Cancer Metastasis Rev. 2016, 35, 235–262. [Google Scholar] [CrossRef] [PubMed]
- Shukla, K.K.; Misra, S.; Sankanagoudar, S.; Sharma, H.; Choudhary, G.R.; Pareek, P.; Vishnoi, J.R.; Sharma, P. Recent scenario of long non-coding RNAs as a diagnostic and prognostic biomarkers of prostate cancer. Urol. Oncol. Semin. Orig. Investig. 2020, 38, 918–928. [Google Scholar] [CrossRef] [PubMed]
- Zijlstra, C.; Stoorvogel, W. Prostasomes as a source of diagnostic biomarkers for prostate cancer. J. Clin. Investig. 2016, 126, 1144–1151. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Li, Y.; Yang, N.; Yang, C.; Mao, L. Current state of biomarkers for the diagnosis and assessment of treatment efficacy of prostate cancer. Discov. Med. 2019, 27, 235–243. [Google Scholar] [PubMed]
- Li, C.; Ye, J.; Zhang, Z.; Gong, Z.; Lin, Z.; Ding, M. Long non-coding RNA RBM5-AS1 promotes the aggressive behaviors of oral squamous cell carcinoma by regulation of miR-1285-3p/YAP1 axis. Biomed. Pharmacother. 2020, 123, 109723. [Google Scholar] [CrossRef] [PubMed]
- Di Cecilia, S.; Zhang, F.; Sancho, A.; De Li, S.; Aguilo, F.; Sun, Y.; Rengasamy, M.; Zhang, W.; Del Vecchio, L.; Salvatore, F.; et al. RBM5-AS1 Is Critical for Self-Renewal of Colon Cancer Stem-like Cells. Cancer Res. 2016, 76, 5615–5627. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Xiao, T.; Ou, W.; Wu, Z.; Wu, J.; Tang, J.; Tian, B.; Zhou, Y.; Su, M.; Wang, W. LncRNA SNHG16 as a potential biomarker and therapeutic target in human cancers. Biomark. Res. 2020, 8, 41. [Google Scholar] [CrossRef] [PubMed]
- Shao, M.; Yu, Z.; Zou, J. LncRNA-SNHG16 Silencing Inhibits Prostate Carcinoma Cell Growth, Downregulate GLUT1 Expression and Reduce Glucose Uptake. Cancer Manag. Res. 2020, 12, 1751–1757. [Google Scholar] [CrossRef] [PubMed]
- Iyer, M.K.; Niknafs, Y.S.; Malik, R.; Singhal, U.; Sahu, A.; Hosono, Y.; Barrette, T.R.; Prensner, J.R.; Evans, J.R.; Zhao, S.; et al. The landscape of long noncoding RNAs in the human transcriptome. Nat. Genet. 2015, 47, 199–208. [Google Scholar] [CrossRef] [PubMed]
- Du, Z.; Fei, T.; Verhaak, R.G.W.; Su, Z.; Zhang, Y.; Brown, M.; Chen, Y.; Liu, X.S. Integrative genomic analyses eveal clinically relevant long noncoding RNAs in human cancer. Nat. Struct. Mol. Biol. 2013, 20, 908–913. [Google Scholar] [CrossRef] [PubMed]
- Malik, B.; Feng, Y.F. Long noncoding RNAs in prostate cancer: overview and clinical implications. Asian J. Androl. 2016, 18, 568–574. [Google Scholar] [PubMed]
- Hua, J.T.; Chen, S.; He, H.H. Landscape of Noncoding RNA in Prostate Cancer. Trends Genet. 2019, 35, 840–851. [Google Scholar] [CrossRef] [PubMed]
- Schmid, M.; Hansen, J.; Chun, F.K.-H. Urinary Prostate Cancer Antigen 3 as a Tumour Marker: Biochemical and Clinical Aspects. In Advances in Cancer Biomarkers, Advances in Experimental Medicine and Biology; Scatena, R., Ed.; Springer Science + Business Media: Dordrecht, The Netherlands, 2015; pp. 227–289. [Google Scholar]
- Prensner, J.R.; Iyer, M.K.; Sahu, A.; Asangani, I.A.; Cao, Q.; Patel, L.R.; Vergara, I.A.; Davicioni, E.; Erho, N.; Ghadessi, M.; et al. The long noncoding RNA SChLAP1 promotes aggressive prostate cancer and antagonizes the SWI/SNF complex. Nat. Genet. 2013, 45, 1392–1398. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.-H.; Ji, J.; Wang, B.-C.; Chen, H.; Yang, Z.-H.; Wang, K.; Luo, C.-L.; Zhang, W.-W.; Wang, F.-B.; Zhang, X.-L. Tumor-Derived Exosomal Long Noncoding RNAs as Promising Diagnostic Biomarkers for Prostate Cancer. Cell. Physiol. Biochem. 2018, 46, 532–545. [Google Scholar] [CrossRef] [PubMed]
- Ren, S.; Wang, F.; Shen, J.; Sun, Y.; Xu, W.; Lu, J.; Wei, M.; Xu, C.; Wu, C.; Zhang, Z.; et al. Long non-coding RNA metastasis associated in lung adenocarcinoma transcript 1 derived miniRNA as a novel plasma-based biomarker for diagnosing prostate cancer. Eur. J. Cancer 2013, 49, 2949–2959. [Google Scholar] [CrossRef] [PubMed]
- Ahadi, A.; Brennan, S.; Kennedy, P.J.; Hutvagner, G.; Tran, N. Long non-coding RNAs harboring miRNA seed regions are enriched in prostate cancer exosomes. Sci. Rep. 2016, 6, 24922. [Google Scholar] [CrossRef] [PubMed]
- Mouraviev, V.; Lee, B.; Patel, V.R.; Albala, D.M.; Johansen, T.E.B.; Partin, A.W.; Ross, A.; Perera, R.J. Clinical prospects of long noncoding RNAs as novel biomarkers and therapeutic targets in prostate cancer. Prostate Cancer Prostat. Dis. 2016, 19, 14–20. [Google Scholar] [CrossRef] [PubMed]
- Crea, F.; Watahiki, A.; Quagliata, L.; Xue, H.; Pikor, L.; Parolia, A.; Wang, Y.; Lin, D.; Lam, W.L.; Farrar, W.L.; et al. Identification of a long non-coding RNA as a novel biomarker and potential therapeutic target for metastatic prostate cancer. Oncotarget 2014, 5, 764–774. [Google Scholar] [CrossRef] [PubMed]
Figure 1.
Distribution of detected lncRNAs across sample types in controls (left) and cases (right).
Figure 1.
Distribution of detected lncRNAs across sample types in controls (left) and cases (right).
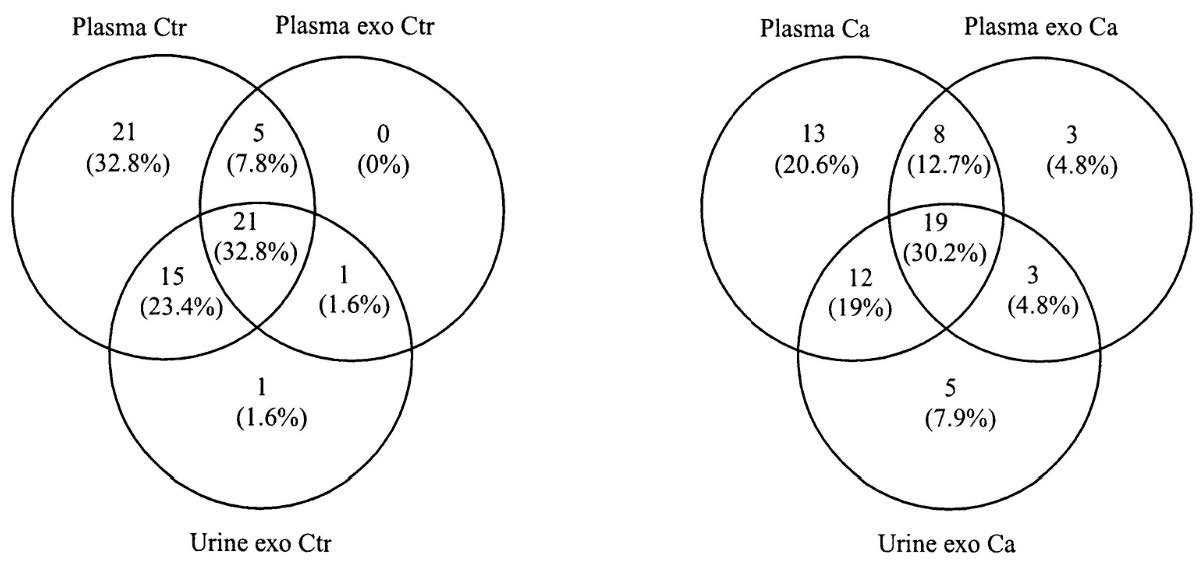
Figure 2.
Volcano plot presenting differentially expressed lncRNAs in plasma of cases versus controls (dotted lines represent the threshold values for p < 0.05 and Fold Change > 2, respectively).
Figure 2.
Volcano plot presenting differentially expressed lncRNAs in plasma of cases versus controls (dotted lines represent the threshold values for p < 0.05 and Fold Change > 2, respectively).
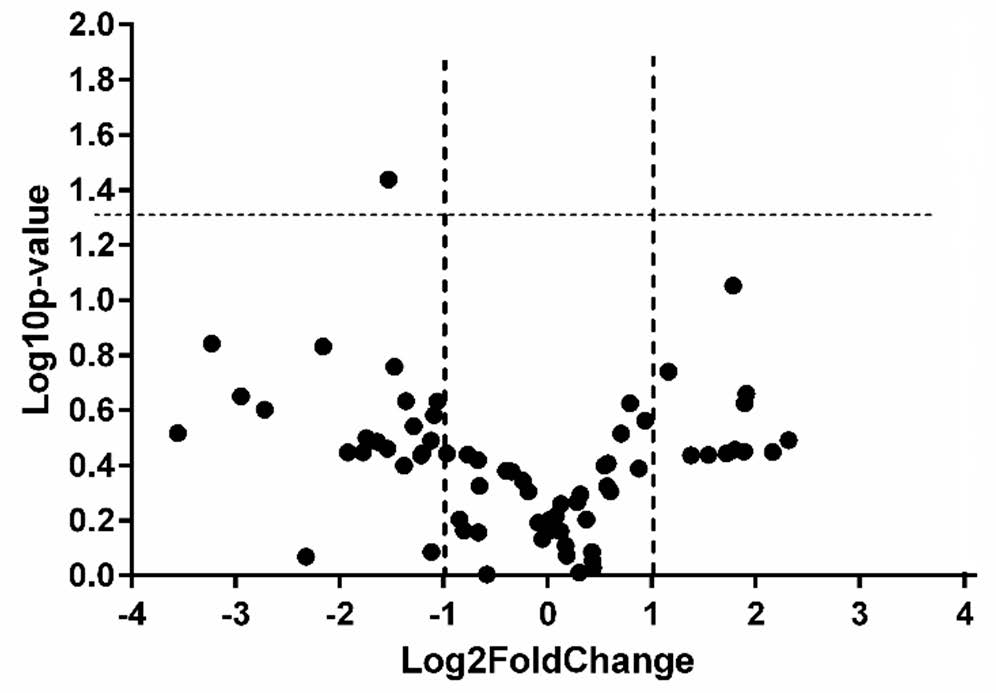
Figure 3.
Volcano plot presenting differentially expressed lncRNAs in urine exosomes of cases versus controls (dotted lines represent the threshold values for p < 0.05 and Fold Change > 2, respectively).
Figure 3.
Volcano plot presenting differentially expressed lncRNAs in urine exosomes of cases versus controls (dotted lines represent the threshold values for p < 0.05 and Fold Change > 2, respectively).
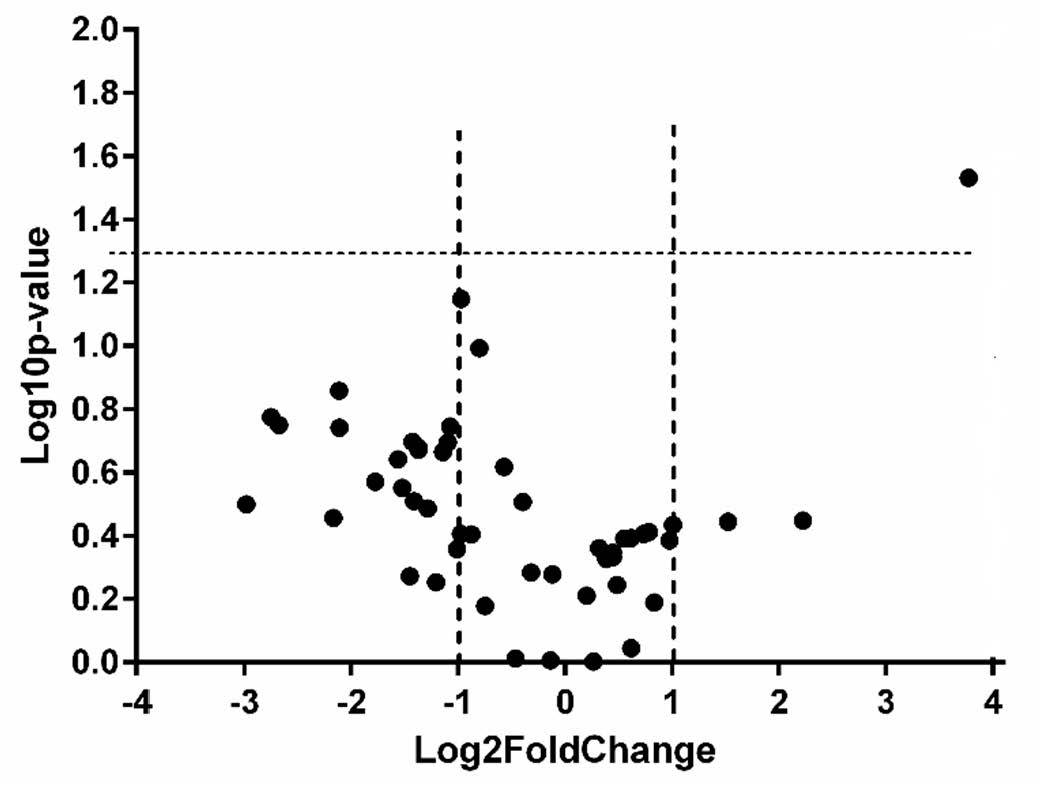
Table 1.
Clinical and demographic characteristics of the study sample.
| Characteristics | Patients (N = 14) | Controls (N = 15) |
|---|---|---|
| Age (years ± SD) | 65.50 (±5.05) | 51.42 (±8.57) |
| PSA | N (%) | N |
| <4 ng/mL | 0 | 15 |
| 4–10 ng/mL | 5 (35.72%) | 0 |
| >10 ng/mL | 9 (64.28%) | 0 |
| Gleason score | N (%) | |
| 5–6 | 2 (14.28) | |
| 7 | 10 (71.43) | |
| 8–10 | 2 (14.28) |
Table 2.
The proportion of detected lncRNAs in the investigated samples.
| Ct Range | Plasma | Plasma Exosomes | Urine Exosomes | |||
|---|---|---|---|---|---|---|
| Patients | Control | Patients | Controls | Patients | Controls | |
| <25 | 6.25 | 6.25 | 6.25 | 6.25 | 6.25 | 6.25 |
| 25–30 | 3.39 | 4.43 | 1.04 | 0 | 2.08 | 1.56 |
| >30 | 15.36 | 19.27 | 10.94 | 10.68 | 13.54 | 12.5 |
| ND | 75.00 | 70.05 | 81.77 | 83.07 | 78.13 | 79.69 |
Note: Data are presented as % detected targets across the entire 96 wells. Ct-cycle threshold. ND-not detected.
Table 3.
Differentially expressed lncRNAs in cases versus controls (*—statistically significant).
| Fold Change Cases vs. Controls | |||
|---|---|---|---|
| lncRNA Gene Symbol | Plasma | Plasma Exosomes | Urine Exosomes |
| AIRN | 3.71 | 2.71 | … |
| ATXN8OS | … | 2.36 | … |
| BANCR | −6.59 | … | … |
| BDNF-AS | 2.24 | … | … |
| BOK-AS1 | −3.42 | … | … |
| CDKN2B-AS1 | … | 7.98 | 4.68 |
| DISC2 | … | … | −3.43 |
| DLX6-AS1 | −2.57 | … | … |
| EGOT | −3.79 | … | … |
| EMX2OS | −2.6 | … | … |
| FALEC | 3.48 | … | … |
| GACAT1 | … | … | −2.59 |
| GAS5 | −5.01 | … | −2.11 |
| H19 | … | −6.41 | |
| HAR1A | −2.18 | −3.06 | −4.32 |
| HEIH | … | −2.76 | … |
| HGDC | −2.22 | … | 4.91 |
| HOTAIR | −2.13 | 4.73 | −2.66 |
| HOTTIP | … | … | 2.01 |
| HOXA11-AS | −2.33 | … | −6.39 |
| HOXA-AS3 | 2.92 | −5.3 | … |
| IPW | −3.11 | … | … |
| KCNIP4-IT1 | 3.71 | … | … |
| KCNQ1OT1 | −11.78 | … | … |
| KRASP1 | … | … | −2.88 |
| LINC00570 | −2.31 | … | … |
| LINC00581 | … | … | 2.87 |
| LINC00853 | −2.09 | … | −2.74 |
| LINC-ROR | −3.35 | 2.09 | −4.49 |
| LUCAT1 | 3.76 | … | … |
| MEG3 | −4.47 | … | … |
| MIAT | 3.29 | … | −2.58 |
| MRPL23-AS1 | … | … | −2.14 |
| NAMA | −2.17 | … | |
| NEAT1 | … | … | −2.69 |
| OIP5-AS1 | −7.73 | 6.17 | … |
| OTX2-AS1 | … | … | −2.95 |
| PCAT1 | 4.98 | … | … |
| PCGEM1 | … | 6.9 | … |
| PRINS | … | … | −6.73 |
| PTCSC1 | … | … | −2.21 |
| PTCSC3 | … | −3.73 | … |
| PTENP1-AS | 3.44 | −2.68 | −2.44 |
| RBM5-AS1 | −2.89 * | … | … |
| RN7SK | … | 5.84 | … |
| RPLP0 | … | −2.19 | … |
| SNHG16 | −9.38 | … | 13.69 * |
| SOX2-OT | … | −3.17 | −2.02 |
| SPRY4-IT1 | −2.91 | −5.74 | … |
| TERC | … | 3.73 | −4.33 |
| TINCR | 2.6 | −2.9 | … |
| TRERNA1 | −2.44 | … | … |
| TUG1 | … | 2.33 | … |
| UCA1 | … | … | −7.89 |
| XIST | 4.48 | … | … |
| ZFAS1 | −2.78 | −2 | −2.31 |
© 2021 Copyright by the authors. Licensed as an open access article using a CC BY 4.0 license.

