Timisoara_Med 2022, 2022(1), 1; doi:10.35995/tmj20220101
Article
SARS-CoV-2 Identification Using Colorimetric Loop-Mediated Isothermal Amplification
1
Department of Biochemistry, “Victor Babes” University of Medicine and Pharmacy, 300041 Timisoara, Romania; paula.muntean22@gmail.com (P.D.M.); moatar.alexandra@umft.ro (A.I.M.); ovidiu.sirbu@umft.ro (I.O.S.)
2
Center for Complex Network Science, “Victor Babes” University of Medicine and Pharmacy, 300041 Timisoara, Romania
3
Department of Pulmonology, “Victor Babes” University of Medicine and Pharmacy, 300041 Timisoara, Romania; oancea@umft.ro
4
Faculty of Medicine, Victor Babes” University of Medicine and Pharmacy, 300041 Timisoara, Romania; octavian.cretu@umft.ro
*
Correspondence: chis.aimee@umft.ro
How to cite: Chiș, A.R.; Muntean, P.D.; Moatăr, A.I.; Oancea, C.; Crețu, O.M.; Sîrbu, I.O. SARS-CoV-2 Identification Using Colorimetric Loop-mediated Isothermal Amplification. Timisoara Med. 2022, 2022(1), 1; doi:10.35995/tmj20220101.
Received: 21 December 2021 / Accepted: 25 January 2022 / Published: 16 February 2022
Abstract
:The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pandemic has caused worldwide losses from all points of view. Although there are efficient vaccines against this virus, rapid detection remains essential for controlling its spread. The gold standard for the COVID-19 diagnostic is the use of quantitative real time polymerase chain reaction (RT-qPCR) from naso-/oro-pharyngeal swabs and, alternatively, saliva specimens. A faster alternative to the RT-qPCR method, the colorimetric reverse transcription loop-mediated isothermal amplification (cLAMP) has a good sensitivity and specificity and has been proposed as an efficient screening method. In this study, we analyzed several sets of LAMP primers for COVID-19 testing using purified RNA samples and raw saliva samples, and tested several reaction conditions. Our results showed that the sensitivity of cLAMP reactions on raw saliva samples can be significantly augmented by the addition of guanidine hydrochloride to levels, which allow its use for COVID-19 screening.
Keywords:
SARS-CoV-2/COVID-19 testing; saliva specimens; cLAMPIntroduction
With over 106 million infections and over two million fatalities, the SARS-CoV-2 pandemic heavily impacted the year 2020 in basically all socio-economic sectors [1,2]. In December 2020, both the US Food and Drug Administration (FDA) and the European Medicines Agency (EMA) approved the Pfizer-BionTech mRNA vaccine for emergency use, and a consistent economic recovery has been witnessed ever since. This was swiftly followed by emergency authorizations of several other vaccines; however, given the continuous appearance of novel SARS-CoV-2 mutated strains, the management of the SARS-CoV-2 pandemic requires the prompt, sensitive and inexpensive detection of any infection to impede further viral spread, controlling the evolution of the pandemic, and the accurate tracing of infected contacts [3,4].
The gold standard for SARS-CoV-2 diagnosis/detection is quantitative real time polymerase chain reaction (RT-qPCR). The method is highly sensitive and specific but requires expensive machines/instruments (real time thermocycler) and well-trained personnel, since the procedure is laborious and the interpretation of the results is not always straight forward. For the time being, the RT-qPCR method is not suitable for large population screening applications [5,6].
Rapid antigen tests are currently widely used as screening tools because they are not time-consuming, and do not require special equipment or highly trained personnel; however, due to their lower sensitivity, they are not the most fortunate alternative to RT-qPCR [4,7].
SARS-CoV-2 infection diagnostics is generally based on the collection of upper (naso- and oro-pharyngeal swabs) and lower (sputum, bronchoalveolar lavage fluid) respiratory tract specimens. The collection of naso- and oro-pharyngeal swabs is unpleasant and requires close contact between the healthcare worker and the patient, increasing the probability of viral contamination. Several research groups have turned their attention towards saliva, which is much easier and safer to collect; furthermore, depending on the disease stage, the saliva viral load could be higher compared to naso-pharyngeal swab specimens [6,8,9,10]. Depending on the method used, the sensitivity of saliva tests varies between 69.2% and 100% [8].
The need for a rapid, sensitive, and specific method has led many research groups to explore the possible use of isothermal amplification, more specifically, the colorimetric loop-mediated isothermal amplification (cLAMP) for SARS-CoV-2 detection [11,12,13,14,15]. The LAMP assay is based on the reverse transcription of SARS-CoV-2 RNA to cDNA followed by isothermal amplification using a set of six primers, and colorimetric monitoring (e.g., using phenol red) of pH changes occurring during target genomic amplification by the Bst 2.0 WarmStart DNA Polymerase. LAMP is highly specific (due to the six primers needed to amplify the target sequence), does not require expensive machines or trained personnel, and can deliver results in 30–60 min [6,16]. The sensitivity of LAMP is comparable to that of RT-qPCR, and has been reported to be even higher for unpurified native samples, which makes it suitable for on-site SARS-CoV-2 testing and screening [6,17,18].
In the present work, we describe our attempt to set up and optimize a LAMP diagnostic method for the detection of SARS-CoV-2 on raw saliva samples. We tested several primers published at the time of performing the experiments and compared their performance with that of LAMP primers of our own design. We show that cLAMP sensitivity could be augmented by the addition of guanidine hydrochloride, while increasing the incubation time or the incubation temperature has deleterious effects.
Materials and Methods
Sample Collection and Processing
Nasopharyngeal swabs and saliva were collected from COVID-19 patients hospitalized at the “Victor Babes” Hospital for Infectious Diseases, Timișoara, Romania. Upon arrival in the lab, all samples were stored at −20 °C until further use, in a fashion which precluded any link to the identity of the patients.
The project was approved by the Ethics Committee of the “Victor Babes” University of Medicine and Pharmacy, Timișoara, Romania. All patients provided written informed consent.
RT-qPCR Confirmation
Viral RNA was purified from naso-pharyngeal swabs on a Maxwell RSC automated Instrument, using a Maxwell® RSC Viral TNA kit (Promega); the viral RNA concentration and purity were estimated using a Nanodrop 1000 spectrophotometer. All samples were then tested for SARS-CoV-2 presence using the GeneFinder Plus RealAmp kit, according to the manufacturer’s protocol. All RT-qPCR runs were validated using both a positive and a negative control.
Saliva Samples Pretreatment
Saliva samples were collected in sterile recipients, centrifuged for 30 s at 1000× g and the supernatant stored at −20 °C until further use; the samples containing food residues and mucus were discarded. All saliva samples were inactivated before processing by adding 40 μL solution of 25 mM NaOH/0.2 mM EDTA solution to 100 μL raw saliva, followed by a brief vortexing and further heat inactivation at 95 °C for 10 min. After cooling on ice, we added 40 μL of 40 mM Tris HCl solution.
Primer Design
The SARS-CoV-2 sequences were retrieved from the NCBI GenBank database (www.ncbi.nlm.nih.gov). Except for Rd-Moh and Na-YZ primers (described by Mohon et al. and Zhang et al.), all the LAMP primers (Rd1, N1, S1, Gapdh) were designed using Primer Explorer V5 and verified by Primer-BLAST (NCI) [19,20]. The schematic representation of LAMP primers is shown in Figure 1. The primer sequences are shown in Table 1.
cLAMP Procedure
cLAMP reactions were performed following the New England BioLabs recommended protocol, using the Warm Start Colorimetric LAMP 2X Master Mix (NEB M1800L). The 25 μL LAMP reaction mix contained 12.5 μL WarmStart Colorimetric LAMP 2X Master Mix, 2.5 μL LAMP Primer Mix (10X) (16 μM Forward Inner Primer—FIP and Backward Inner Primer—BIP, 2 μM F3 and B3, 4 μM Loop Forward—LF and Loop Backward LB), 1 μL target RNA and 9 μL RNase/DNase free H2O. LAMP mixtures were incubated at 65 °C for 30–45 min in a closed lid Water Bath/Thermoblock. Sample photographs were taken using an iPhone XS cellular phone camera. Samples which turned color from pink (the original color of phenol red) to yellow were considered positive (Figure 2A).
We tested several reaction conditions: temperatures (65 and 70 °C) and incubation time (30 and 45 min); furthermore, we attempted to optimize the procedure by adding 3 μL (5 μM final concentration) or 6 μL (10 μM final concentration) of guanidine hydrochloride 40 mM.
The LAMP amplification products were visualized by gel electrophoresis on 2% agarose gel stained with ethidium bromide (Figure 2B).
Calculations
Sensitivity (true positive rate) was calculated as the ratio
Specificity (true negative rate) was calculated as the ratio .
All statistical tests were performed using Prism 9.2.0 for Mac.
Results
RT-qPCR Confirmation Using the GeneFinder
We used the GeneFinder RT-qPCR kit to confirm the presence of SARS-CoV-2 in 33 naso-pharyngeal swab samples. The GeneFinder kit tests were used to identify the presence of the RdRp, N and E genes in the SARS-CoV-2 genome. The amplification characteristics for the three genes are presented in Table 2.
A one-way ANOVA Tukey multiple comparisons test showed that there were no statistically significant differences between the three data sets, with adjusted p values of 0.998 (RdRp vs. N gene), 0.9965 (N gene vs. E gene) and 0.994 (RdRp vs. E gene). Furthermore, the Pearson correlation analysis showed a very good correlation between RdRp, N and E genes (r > 0.92, p < 0.0001) in the RT-qPCR assay. Altogether, these data suggest that, in the context of the RT-qPCR assay, all three genes were equally suitable to detect the presence of SARS-CoV-2 in the naso-pharyngeal samples.
Colorimetric LAMP on Purified Viral RNA
First, we tested the Rd-Moh-4 and Na-YZ primers on the same 33 positive samples and on additional 30 negative viral RNA samples. After 30 min of incubation at 65 °C, the calculated sensitivities of Rd-Moh-4 and Na-YZ primers were 93% and 97% respectively; further incubation to 45 min did not alter these results. Specificity calculations for Rd-Moh-4 showed 88% at 30 min, with a surprising decrease to 73% at 45 min; unexpectedly, for Na-YZ primers, all negative samples turned positive, therefore, specificity was 0 (Table 3). An overall analysis of the Ct value correlation with the cLMAP assay results indicated a surprisingly modest (r = 0.623) statistically significant correlation (p = 2.15 × 10−0.3) only for the RdRp gene.
Next, we tested primers of our own design (Rd1, N1 and S1) on another set of 30 samples. Rd1 showed a low sensitivity, 39% at 30 min and 42% at 45 min, while specificity reached 100% (both at 30 and 45 min). S1 sensitivity was 58% at 30 min and 64% at 45 min, with a specificity of 80% at 30 min and 83% at 45 min. Of note, four negative samples turned an unusual orange color at 45 min; however, the gel electrophoresis analysis showed they are trully negative results. Intriguingly, similar to the Na-YZ primers, N1 showed 100% sensitivity but 0% specificity (Table 3), most probably due to an unspecific polymerization. Since in our previous RT-qPCR experiment, only RdRp was correlated to the cLAMP results, we focused on RdRp amplification as a control method for cLAMP. RdRp Ct values were significantly and strongly correlated with Rd1 (r = −0.898; p = 1.8 × 10−11 and r = −0.916, p = 1.24 × 10−12 at 30 and 45 min, respectively). Correlation coefficients were more modest for S1 primers: r = −0.73; p = 4.6 × 10−6; and r = −0.69, p = 2.1 × 10−5.
Of note, all (positive and negative) samples turned yellow for Gapdh primers, indicating that the enzyme is functional under the specific conditions of our protocol.
Colorimetric LAMP on Raw Saliva Samples
Next, we investigated whether the LAMP primers may be used to detect the presence of SARS-CoV-2 in saliva samples. Unexpectedly, in the cLMAP on saliva setting, Rd-Moh showed zero specificity (data not shown) and was further excluded from our analysis.
The use of Rd1 and S1 primers on 60 positive saliva samples yielded a sensitivity of 17% and 80%, respectively, at 30 min incubation at 65 °C. The analysis of 20 negative saliva samples showed a 100% specificity for Rd1 and 75% for S1. Increasing the incubation time to 45 min had no effect on sensitivity, while S1 specificity unexpectedly dropped to 35% (Table 4). All samples turned positive when tested with the Gapdh control. The results were reproducible over a two-week period of time, with saliva samples stored aliquoted at −20 °C. Of note, no information regarding the qRT-PCR kit used (or the Ct values) for the COVID-19 RT-qPCR diagnostic in naso-pharyngeal swabs could be retrieved. Furthermore, no information regarding the time of saliva collection relative to the RT-qPCR diagnostic time was provided.
Colorimetric LAMP Testing in the Presence of Guanidine Hydrochloride
Since Rd1 showed an excellent specificity, we tried to increase its sensitivity by supplementing the LAMP reaction mix with guanidine hydrochloride at final concentrations of 5 μM and 10 μM [21].
In a different set of 20 positive saliva samples, 5 μM and 10 μM guanidine hydrochloride significantly augmented the Rd1 sensitivity at 30 min from an initial 15% to 55% and 90%, respectively. The testing of 20 negative saliva samples showed a specificity of 100% for 5 μM guanidine hydrochloride and 85% for 10 μM guanidine hydrochloride. Of note, the specificity for 5 μM and 10 μM guanidine hydrochloride reaction mix dropped to 70% and 40%, respectively, when the incubation time was extended to 45 min (Table 5). Furthermore, an increase in the incubation temperature to 70 °C decreased the sensitivity, regardless of the time of incubation (data not shown).
Discussion
The LAMP assay has multiple advantages compared to the RT-PCR method: it eliminates the need for a thermocycler (since the amplification takes place at a constant temperature), and the target amplification can be monitored by using pH sensitive dyes such as phenol red (since the addition of each nucleotide to the nascent strand liberates a proton). Here, we showed that the direct detection of SARS-CoV-2 in native, raw saliva samples by cLAMP is feasible even in suboptimal conditions, and its sensitivity can be significantly improved by the addition of guanidinium hydrochloride.
The sensitivity of LAMP assay has been shown to be lower compared to the RT-qPCR assay, irrespective of its use on raw or purified samples [15]. The reports on LAMP assay show widely variable performances; in this respect, the performance of our cLAMP method of analysis of saliva samples is in line with data communicated by others [8,22,23]. Pasomsub et al. reported an 84.2% sensitivity and 98.9% specificity for a qRT-PCR-based analysis of saliva samples, using nasopharyngeal swabs as a reference [24]. An even higher sensitivity was noted by To et al. and Azzi et al.: 91.7% and 100% of the saliva samples tested were positive, respectively, while Becker et al. communicated the lowest sensitivity (69.2%) [25].
The efficiency of cLAMP tests based on monitoring the change in pH during target amplification is influenced by the level of SRAR-CoV2 load in the sample. Dao Thi et al. reported that only 10% of the cLAMP reactions turned positive on RNA samples with Ct between 30 and 35. However, this cannot explain the differences in cLAMP sensitivities observed in tests performed on purified RNA samples, since the naso-pharyngeal swabs analyzed showed an average Ct of around 26 for all three genes (RdRp, N and E).
Since a LAMP assay should be able to detect as low as two copies of SARS-CoV-2 RNA, we are tempted to speculate that the differences in sensitivity reflect the impact of improper RNA sample handling (repeated thawing/freezing cycles, contamination with nucleases, etc.) [14].
Our saliva pretreatment protocol included 10 min heat treatment at 95 °C; heat is known to inactivate the virus, protect its RNA from degradation (by inactivating nucleases) and to improve SARS-CoV-2 detection in LAMP assays [26]. Furthermore, together with the detergents and NaOH used, the heat contributes to the denaturation of viral proteins and promotes the access of Bst2 to the viral genome.
Multiple additives (DMSO, polyethylene glycol, betaine and guanidinium hydrochloride) have been tested for their ability to improve the performance of cLAMP [21]. In our raw saliva experiments, the addition of guanidium hydrochloride to a final concentration of 10 μM significantly improved sensitivity, possibly through the denaturation of salivary ribonuclease inhibitors, and enhanced the base pairing of the primers with the target DNA strand [21]. However, we were not able to see a lower detection time, as previously reported by Zhang et al. (2020), or improvements in specificity (data not shown).
The sensitivity and specificity of our cLAMP assay on saliva samples is obviously inferior to that of the RT-qPCR method on nasopharyngeal swabs. Nevertheless, at 80% overall sensitivity and specificity, our method compensates for this due to its rapid turnaround and simplicity. It has been shown that saliva cLAMP assays turn positive at rather high viral loads (>102 viral copies/µL) (Kobayashi et al., 2021) [27]. However, in a clinical, epidemiologically relevant setting, it is important to identify the individuals with high transmissibility potential, which correlates with higher viral loads [28]. Furthermore, the efficiency of a surveillance protocol depends on the frequency and the turnaround time of the test used, rather than on its sensitivity [29].
The importance of salivary testing is underlined by the recent observation that saliva viral load correlates with clinical severity [30]. The salivary viral load is maximal in the first two days of the symptomatic period and declines steadily (together with transmissibility) in the next seven to nine days; this suggests that salivary testing should best be used as s screening method, especially at the very beginning of the symptomatic period [31].
Due to the limited number of biological samples included in our study, we could not stratify the cohorts and analyze the influence of other factors such as the age, sex, drinking and smoking habits, and severity of disease. Age and gender have been shown to significantly influence viral shedding and clearance; this would explain the reported differences in testing results, with older males more often testing positive after the 5th day of disease [32,33]. Patients’ compliance is another issue which has negatively impacted our study; not only were the patients reluctant to provide saliva samples, but a significant number of these samples contained food contaminants, despite our specific request for a minimum of 60 min fasting before collection. Food contaminants might contain polymerase inhibitors, while food decay could lead to significant changes in pH and thus significantly impact the final result [34].
In conclusion, the isothermal 10 μM guanidinium chloride spiked cLAMP procedure on raw saliva samples could be taken into consideration as a reliable self-testing procedure, since its ease of use clearly compensates for the lower sensitivity and specificity.
Author Contributions
Conceptualization, A.R.C. and I.O.S.; Methodology, A.R.C. and P.D.M.; Formal Analysis, A.R.C., A.I.M. and P.D.M.; Resources, O.M.C. and C.O.; Data Curation, I.O.S.; Writing—Original Draft Preparation, A.R.C.; Writing—Review and Editing, A.R.C. and I.O.S.; Visualization, A.I.M.; Supervision, I.O.S.; Project Administration, A.R.C.; Funding Acquisition, O.M.C. and C.O.
Funding
This research received no external funding.
Acknowledgments
We are grateful to Larisa Stefanut-Turcut for the technical support.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Baker, S.R.; Bloom, N.; Davis, S.J.; Terry, S.J. Covid-Induced Economic Uncertainty; Working Paper 26983; NBER: Cambridge, MA, USA, 2020. [Google Scholar]
- Hevia, C.; Neumeyer, A. A Conceptual Framework for Analyzing the Economic Impact of COVID-19 and Its Policy Implications; UNDP LAC COVID-19 Policy Documents Series; United Nations Development Programme: New York, NY, USA, 2020; pp. 1–29. [Google Scholar]
- Kashir, J.; Yaqinuddin, A. Loop mediated isothermal amplification (LAMP) assays as a rapid diagnostic for COVID-19. Med. Hypotheses 2020, 141, 109786. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Xie, T.; Tong, Y. Rapid and highly sensitive one-tube colorimetric RT-LAMP assay for visual detection of SARS-CoV-2 RNA. Biosens. Bioelectron 2021, 187, 113330. [Google Scholar] [CrossRef] [PubMed]
- Chow, F.W.; Chan, T.T.; Tam, A.R.; Zhao, S.; Yao, W.; Fung, J.; Cheng, F.K.; Lo, G.C.; Chu, S.; Aw-Yong, K.L.; et al. A rapid, simple, inexpensive, and mobile colorimetric assay COVID-19-LAMP for mass on-site screening of COVID-19. Int. J. Mol. Sci. 2020, 21, 5380. [Google Scholar] [CrossRef] [PubMed]
- Lalli, M.A.; Langmade, J.S.; Chen, X.; Fronick, C.C.; Sawyer, C.S.; Burcea, L.C.; Wilkinson, M.N.; Fulton, R.S.; Heinz, M.; Buchser, W.J.; et al. Rapid and extraction-free detection of SARS-CoV-2 from saliva by colorimetric reverse-transcription loop-mediated isothermal amplification. Clin. Chem. 2021, 67, 415–424. [Google Scholar] [CrossRef]
- Scohy, A.; Anantharajah, A.; Bodéus, M.; Kabamba-Mukadi, B.; Verroken, A.; Rodriguez-Villalobos, H. Low performance of rapid antigen detection test as frontline testing for COVID-19 diagnosis. J. Clin. Virol. 2020, 129, 104455. [Google Scholar] [CrossRef]
- To, K.K.; Tsang, O.T.; Yip, C.C.; Chan, K.H.; Wu, T.C.; Chan, J.M.; Leung, W.S.; Chik, T.S.; Choi, C.Y.; Kandamby, D.H.; et al. Consistent detection of 2019 novel coronavirus in saliva. Clin. Infect. Dis. 2020, 71, 841–843. [Google Scholar] [CrossRef]
- Wyllie, A.L.; Fournier, J.; Casanovas-Massana, A.; Campbell, M.; Tokuyama, M.; Vijayakumar, P.; Warren, J.L.; Geng, B.; Muenker, M.C.; Moore, A.J.; et al. Saliva or nasopharyngeal swab specimens for detection of SARS-CoV-2. NEJM 2020, 383, 1283–1286. [Google Scholar] [CrossRef]
- Xun, G.; Lane, S.T.; Petrov, V.A.; Pepa, B.E.; Zhao, H. A rapid, accurate, scalable, and portable testing system for COVID-19 diagnosis. Nat. Commun. 2021, 12, 2905. [Google Scholar] [CrossRef]
- Lamb, L.E.; Bartolone, S.N.; Ward, E.; Chancellor, M.B. Rapid detection of novel coronavirus/Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) by reverse transcription-loop-mediated isothermal amplification. PLoS ONE 2020, 15, e0234682. [Google Scholar] [CrossRef]
- Rabe, B.A.; Cepko, C. SARS-CoV-2 detection using isothermal amplification and a rapid, inexpensive protocol for sample inactivation and purification. Proc. Natl. Acad. Sci. USA 2020, 117, 24450–23358. [Google Scholar] [CrossRef]
- Österdahl, M.F.; Lee, K.A.; Lochlainn, M.N.; Wilson, S.; Douthwaite, S.; Horsfall, R.; Sheedy, A.; Goldenberg, S.D.; Stanley, C.J.; Spector, T.D.; et al. Detecting SARS-CoV-2 at point of care: Preliminary data comparing loop-mediated isothermal amplification (LAMP) to polymerase chain reaction (PCR). BMC Infect. Dis. 2020, 20, 783. [Google Scholar] [CrossRef]
- Huang, W.E.; Lim, B.; Hsu, C.C.; Xiong, D.; Wu, W.; Yu, Y.; Jia, H.; Wang, Y.; Zeng, Y.; Ji, M.; et al. RT-LAMP for rapid diagnosis of coronavirus SARS-CoV-2. Microb. Biotechnol. 2020, 13, 950–961. [Google Scholar] [CrossRef]
- Thi, V.L.; Herbst, K.; Boerner, K.; Meurer, M.; Kremer, L.P.; Kirrmaier, D.; Freistaedter, A.; Papagiannidis, D.; Galmozzi, C.; Stanifer, M.L.; et al. A colorimetric RT-LAMP assay and LAMP-sequencing for detecting SARS-CoV-2 RNA in clinical samples. Sci. Transl. Med 2020, 12, eabc7075. [Google Scholar]
- Notomi, T.; Okayama, H.; Masubuchi, H.; Yonekawa, T.; Watanabe, K.; Amino, N.; Hase, T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000, 28, e63. [Google Scholar] [CrossRef]
- Kaneko, H.; Kawana, T.; Fukushima, E.; Suzutani, T. Tolerance of loop-mediated isothermal amplification to a culture medium and biological substances. J. Biochem. Biophys. Methods 2007, 70, 499–501. [Google Scholar] [CrossRef]
- Francois, P.; Tangomo, M.; Hibbs, J.; Bonetti, E.J.; Boehme, C.C.; Notomi, T.; Perkins, M.D.; Schrenzel, J. Robustness of a loop-mediated isothermal amplification reaction for diagnostic applications. FEMS Immunol. Med. Microbiol. 2011, 62, 41–48. [Google Scholar] [CrossRef]
- Mohon, A.N.; Hundt, J.; van Marle, G.; Pabbaraju, K.; Berenger, B.; Griener, T.; Lisboa, L.; Church, D.; Czub, M.; Greninger, A.; et al. Development and validation of direct RT-LAMP for SARS-CoV-2. medRxiv 2020. [Google Scholar] [CrossRef]
- Zhang, Y.; Odiwuor, N.; Xiong, J.; Sun, L.; Nyaruaba, R.O.; Wei, H.; Tanner, N.A. Rapid Molecular Detection of SARS-CoV-2 (COVID-19) Virus RNA Using Colorimetric LAMP. medRxiv 2020. [Google Scholar] [CrossRef]
- Zhang, Y.; Ren, G.; Buss, J.; Barry, A.J.; Patton, G.C.; Tanner, N.A. Enhancing colorimetric loop-mediated isothermal amplification speed and sensitivity with guanidine chloride. Biotechniques 2020, 69, 178–185. [Google Scholar] [CrossRef]
- Azzi, L.; Carcano, G.; Gianfagna, F.; Grossi, P.; Dalla Gasperina, D.; Genoni, A.; Fasano, M.; Sessa, F.; Tettamanti, L.; Carinci, F.; et al. Saliva is a reliable tool to detect SARS-CoV-2. J. Infect. 2020, 81, e45–e50. [Google Scholar] [CrossRef]
- To, K.K.; Tsang, O.T.; Leung, W.S.; Tam, A.R.; Wu, T.C.; Lung, D.C.; Yip, C.C.; Cai, J.P.; Chan, J.M.; Chik, T.S.; et al. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: An observational cohort study. Lancet Infect. Dis. 2020, 20, 565–574. [Google Scholar] [CrossRef]
- Pasomsub, E.; Watcharananan, S.P.; Boonyawat, K.; Janchompoo, P.; Wongtabtim, G.; Suksuwan, W.; Sungkanuparph, S.; Phuphuakrat, A. Saliva sample as a non-invasive specimen for the diagnosis of coronavirus disease 2019: A cross-sectional study. Clin. Microbiol. Infect. 2021, 27, 285.e1–285.e4. [Google Scholar] [CrossRef] [PubMed]
- Becker, D.; Sandoval, E.; Amin, A.; De Hoff, P.; Leonetti, N.; Lim, Y.W.; Elliott, C.; Laurent, L.; Grzymski, J.; Lu, J. Saliva is less sensitive than nasopharyngeal swabs for COVID-19 detection in the community setting. medRxiv 2020. [Google Scholar] [CrossRef]
- Burton, J.; Love, H.; Richards, K.; Burton, C.; Summers, S.; Pitman, J.; Easterbrook, L.; Davies, K.; Spencer, P.; Killip, M.; et al. The effect of heat-treatment on SARS-CoV-2 viability and detection. J. Virol. Methods 2021, 290, 114087. [Google Scholar] [CrossRef]
- Kobayashi, G.S.; Brito, L.A.; Moreira, D.D.; Suzuki, A.M.; Hsia, G.S.; Pimentel, L.F.; de Paiva, A.P.; Dias, C.R.; Lourenço, N.C.; Oliveira, B.A.; et al. A Novel Saliva RT-LAMP Workflow for Rapid Identification of COVID-19 Cases and Restraining Viral Spread. Diagnostics 2021, 11, 1400. [Google Scholar] [CrossRef]
- Marks, M.; Millat-Martinez, P.; Ouchi, D.; Roberts, C.H.; Alemany, A.; Corbacho-Monné, M.; Ubals, M.; Tobias, A.; Tebé, C.; Ballana, E.; et al. Transmission of COVID-19 in 282 clusters in Catalonia, Spain: A cohort study. Lancet Infect. Dis. 2021, 21, 629–636. [Google Scholar] [CrossRef]
- Larremore, D.B.; Wilder, B.; Lester, E.; Shehata, S.; Burke, J.M.; Hay, J.A.; Tambe, M.; Mina, M.J.; Parker, R. Test sensitivity is secondary to frequency and turnaround time for COVID-19 screening. Sci. Adv. 2021, 7, eabd5393. [Google Scholar] [CrossRef]
- Silva, J.; Lucas, C.; Sundaram, M.; Israelow, B.; Wong, P.; Klein, J. Saliva viral load is a dynamic unifying correlate of COVID-19 severity and mortality. medRxiv 2021. [Google Scholar] [CrossRef]
- Hu, S.; Wang, W.; Wang, Y.; Litvinova, M.; Luo, K.; Ren, L. Infectivity, susceptibility, and risk factors associated with SARS-CoV-2 transmission under intensive contact tracing in Hunan, China. Nat. Commun. 2021, 12, 1533. [Google Scholar] [CrossRef]
- Takahashi, T.; Ellingson, M.K.; Wong, P.; Israelow, B.; Lucas, C.; Klein, J.; Silva, J.; Mao, T.; Oh, J.E.; Tokuyama, M.; et al. Sex differences in immune responses that underlie COVID-19 disease outcomes. Nature 2020, 588, 315–320. [Google Scholar] [CrossRef]
- Zhou, C.; Zhang, T.; Ren, H.; Sun, S.; Yu, X.; Sheng, J.; Shi, Y.; Zhao, H. Impact of age on duration of viral RNA shedding in patients with COVID-19. Aging 2020, 12, 22399–22404. [Google Scholar] [CrossRef]
- Uribe-Alvarez, C.; Lam, Q.; Baldwin, D.A.; Chernoff, J. Low saliva pH can yield false positives results in simple RT-LAMP-based SARS-CoV-2 diagnostic tests. PLoS ONE 2021, 16, e0250202. [Google Scholar] [CrossRef]
Figure 1.
Schematic representation of the RT-LAMP primers.
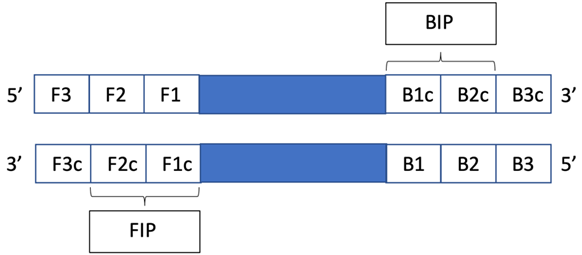
Figure 2.
Typical colorimetric and electrophoretic LAMP results for RdRp primers. (A) cLAMP results showing LAMP amplification (1–3) with specific change in color from pink to yellow. (B) Electrophoforetic aspect of RdRp amplicon migration in electric field: 5, 10: positive samples; 22: negative sample; H2O: negative control 1 (distilled water); RNA: negative control 2 (mouse RNA).
Figure 2.
Typical colorimetric and electrophoretic LAMP results for RdRp primers. (A) cLAMP results showing LAMP amplification (1–3) with specific change in color from pink to yellow. (B) Electrophoforetic aspect of RdRp amplicon migration in electric field: 5, 10: positive samples; 22: negative sample; H2O: negative control 1 (distilled water); RNA: negative control 2 (mouse RNA).
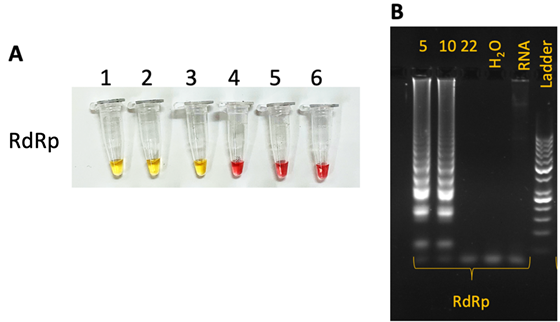
Table 1.
Sequences of the primers used.
| Label | Sequence 5′-3′ |
| Rd1-F3 | ACAAAGCCTTACATTAAGTGG |
| Rd1-B3 | CACCATCAACAAATATTTTTCTCAC |
| Rd1-FIP | TGGGTGGTATGTCTGATCCCAATAGATTTGTTAAAATATGACTTCACGG |
| Rd1-BIP | TGTGTTAACTGTTTGGATGACAGATGTAGGTGGGAACACTGT |
| Rd1-LF | CGGTCAAAGAGTTTTAACCTCTCTT |
| Rd1-LB | TGCATTCTGCATTGTGCAAACT |
| Label | Sequence 5′-3′ |
| N1-F3 | TGGCTACTACCGAAGAGCT |
| N1-B3 | TGCAGCATTGTTAGCAGGAT |
| N1-FIP | TCTGGCCCAGTTCCTAGGTAGTGACGAATTCGTGGTGGTGA |
| N1-BIP | AGACGGCATCATATGGGTTGCAGCGGGTGCCAATGTGATC |
| N1-LF | ACCATCTTGGACTGAGATCTTTCAT |
| N1-LB | ACTGAGGGAGCCTTGAATACACC |
| Label | Sequence 5′-3′ |
| Rd-Moh-4-F4 | ACATGCTTAGAATTATGGCC |
| Rd-Moh-4-B4 | GCTTGACAAATGTTAAAAACACT |
| Rd-Moh-4-FIP | TTGAGCACACTCATTAGCTAATCTATCACTTGTTCTTGCTCGCA |
| Rd-Moh-4-BIP | GAGTGAAATGGTCATGTGTGGCGCATAAGCAGTTGTGGCA |
| Rd-Moh-4-LPF | GACAAGCTACAACACGTTGTATGT |
| Rd-Moh-4-LPB | ACTATATGTTAAACCAGGTGGAACC |
| Label | Sequence 5′-3′ |
| Na-YZ-F3 | TGGCTACTACCGAAGAGCT |
| Na-YZ-B3 | TGCAGCATTGTTAGCAGGAT |
| Na-YZ-FIP | TCTGGCCCAGTTCCTAGGTAGTCCAGACGAATTCGTGGTGG |
| Na-YZ-BIP | AGACGGCATCATATGGGTTGCACGGGTGCCAATGTGATCT |
| Na-YZ-LF | GGACTGAGATCTTTCATTTTACCGT |
| Na-YZ-LB | ACTGAGGGAGCCTTGAATACA |
| Label | Sequence 5′-3′ |
| S1-F3 | GGTGTTTATTACCCTGACAAAG |
| S1-B3 | GTACCAAAAATCCAGCCTC |
| S1-FIP | CATGGAACCAAGTAACATTGGAAAATTTTCAGATCCTCAGTTTTACATTC |
| S1-BIP | CTCTGGGACCAATGGTACTAAGAGGACTTCTCAGTGGAAGCA |
| S1-LF | GAAAGGTAAGAACAAGTCCTGAGT |
| S1-LB | CTGTCCTACCATTTAATGATGGTGT |
| Label | Sequence 5′-3′ |
| Gapdh-F3 | AGAACGGGAAGCTTGTCATC |
| Gapdh-B3 | CGAACATGGGGGCATCAG |
| Gapdh-FIP | GACGTACTCAGCGCCAGCATATCTTCCAGGAGCGAGATCC |
| Gapdh-BIP | GCGTCTTCACCACCATGGAGACAGAGGGGGCAGAGATGA |
| Gapdh-LF | CGCCCCACTTGATTTTGGA |
| Gapdh-LB | TGCAGGGGGGAGCCAAAAG |
Table 2.
Cycle threshold (Ct) characteristics for the RdRp, N and E amplification curves.
| RdRp | E | N | |
|---|---|---|---|
| Average Ct | 26.29 | 26.67 | 26.09 |
| Standard Deviation | 4.031 | 4.325 | 3.509 |
| Variation Coefficient (%) | 15.34 | 16.22 | 13.45 |
| Median | 26.32 | 26.33 | 26.05 |
| Min/Max | 14.74/36.24 | 21.24/41.91 | 20.80/37.54 |
Table 3.
Overall performance of LAMP assays on RNA extracted from nasopharyngeal swabs from positive and negative patients.
Table 3.
Overall performance of LAMP assays on RNA extracted from nasopharyngeal swabs from positive and negative patients.
| Time (min) | 30 | 45 | ||||||||||
| LAMP primer | Rd-Moh-4 | Na-YZ | Rd1 | N1 | S1 | Gapdh | Rd-Moh-4 | Na-YZ | Rd1 | N1 | S1 | Gapdh |
| Positive RNA samples | 30 | 30 | 30 | 30 | ||||||||
| cLAMP positive result | 28 | 29 | 13 | 30 | 19 | 33 | 28 | 29 | 14 | 30 | 21 | 30 |
| Sensitivity % | 93 | 97 | 43.3 | 100 | 58 | 100 | 93.3 | 96.6 | 46.7 | 100 | 70 | 100 |
| Negative RNA samples | 41 | 30 | 41 | 30 | ||||||||
| cLAMP negative result | 36 | 0 | 30 | 0 | 25 | 30 | 30 | 0 | 30 | 0 | 21 | 30 |
| Specificity % | 88 | 0 | 100 | 0 | 83 | 100 | 73 | 0 | 100 | 0 | 70 | 100 |
Table 4.
Overall performance of LAMP assays for (positive and negative) raw saliva samples for Rd1 and S1 primers at 65 °C.
Table 4.
Overall performance of LAMP assays for (positive and negative) raw saliva samples for Rd1 and S1 primers at 65 °C.
| Time (min) | 30 | 45 | ||||
| LAMP primer | Rd1 | S1 | Gapdh | Rd1 | S1 | Gapdh |
| Positive RNA samples | 60 | |||||
| cLAMP positive result | 10 | 48 | 60 | 11 | 48 | 60 |
| Sensitivity % | 17 | 80 | 100 | 18 | 80 | 100 |
| Negative RNA samples | 20 | |||||
| cLAMP negative result | 20 | 15 | 20 | 20 | 7 | 20 |
| Specificity % | 100 | 75 | 100 | 100 | 35 | 100 |
Table 5.
Overall performance of LAMP assays for (positive and negative) saliva samples for Rd1 and S1 primers at 65 °C, in the presence of guanidine.
Table 5.
Overall performance of LAMP assays for (positive and negative) saliva samples for Rd1 and S1 primers at 65 °C, in the presence of guanidine.
| Time (min) | 30 | 45 | ||||
| Rd1 primer | no guanidine HCl | 5 μM guanidine HCl | 10 μM guanidine HCl | no guanidine HCl | 5 μM guanidine HCl | 10 μM guanidine HCl |
| Positive RNA samples | 20 | |||||
| Result positive samples | 3 | 11 | 16 | 3 | 11 | 16 |
| Sensitivity % | 15 | 55 | 80 | 15 | 55 | 80 |
| Negative RNA samples | 20 | |||||
| Result negative samples | 20 | 20 | 18 | 20 | 14 | 8 |
| Specificity % | 100 | 100 | 90 | 100 | 70 | 40 |
© 2022 Copyright by the authors. Licensed as an open access article using a CC BY 4.0 license

